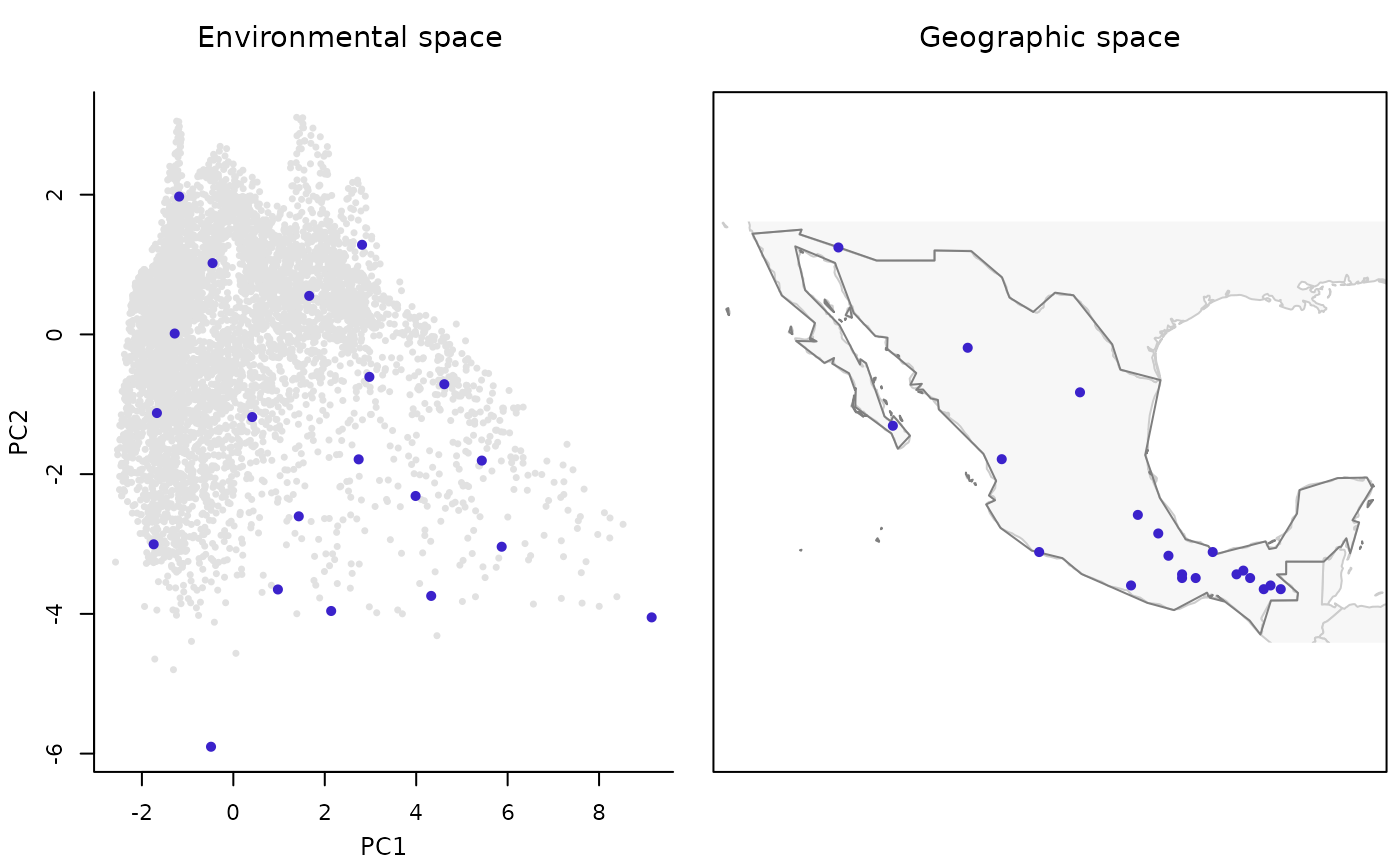
Plots representing sites (all and selected for survey) in environmental and/or geographic space.
Usage
plot_sites_EG(master_selection, selection_type, variable_1 = NULL,
variable_2 = NULL, selection_number = 1,
region_border = TRUE, mask_border = FALSE, col_all = NULL,
col_sites = NULL, col_pre = NULL, cex_all = 0.7,
cex_sites = 1, cex_pre = 1, pch_all = 16, pch_sites = 16,
pch_pre = 16, add_main = TRUE, mar = NULL)
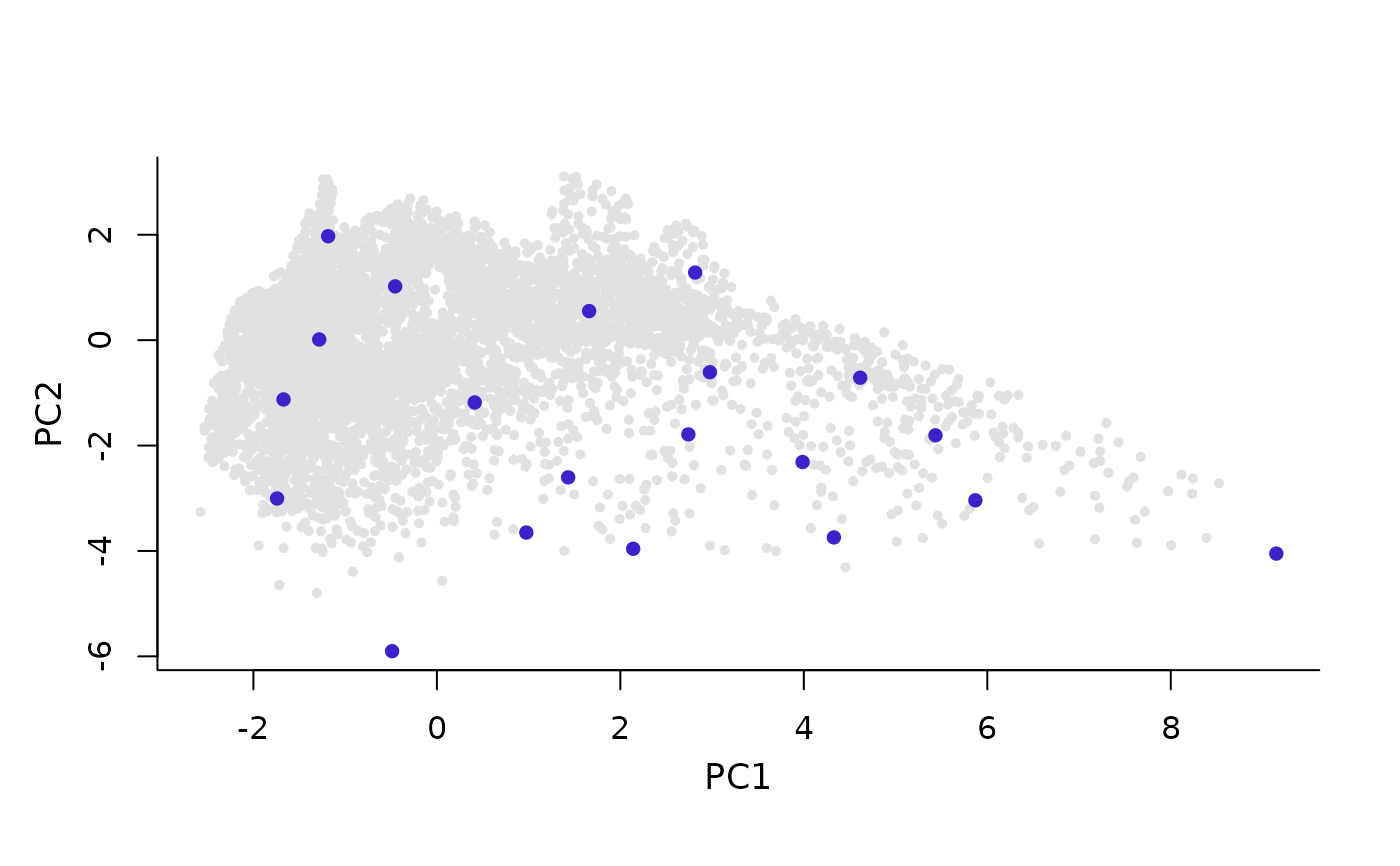
plot_sites_E(master_selection, selection_type, variable_1 = NULL,
variable_2 = NULL, selection_number = 1, col_all = NULL,
col_sites = NULL, col_pre = NULL, cex_all = 0.7,
cex_sites = 1, cex_pre = 1, pch_all = 16, pch_sites = 16,
pch_pre = 16, main = "", xlab = NULL, ylab = NULL)
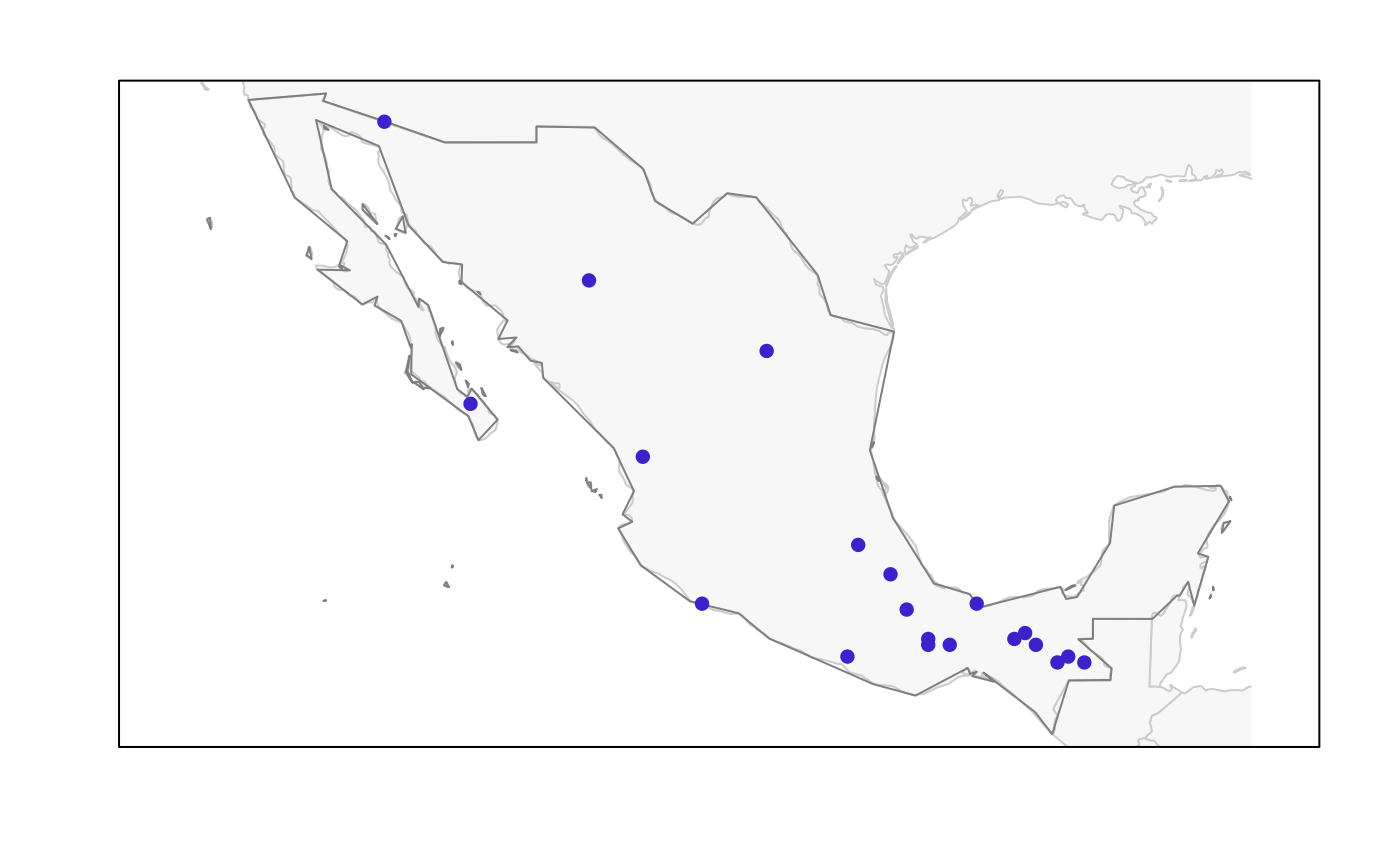
plot_sites_G(master_selection, selection_type, selection_number = 1,
region_border = TRUE, mask_border = FALSE, col_all = NULL,
col_sites = NULL, col_pre = NULL, cex_all = 0.7,
cex_sites = 1, cex_pre = 1, pch_all = 16, pch_sites = 16,
pch_pre = 16, mar = NULL)Arguments
- master_selection
master_selection object derived from functions
random_selection,uniformG_selection,uniformE_selection, orEG_selection.- selection_type
(character) type of selection depending on the function used to select sites. The options available are "random" (
random_selection), "G" (uniformG_selection), "E" (uniformE_selection), and "EG" (EG_selection).- variable_1
(character or numeric) name or position of the first variable (x-axis) to be plotted in environmental space. Default = NULL, required when
selection_type= "random" or "G".- variable_2
(character or numeric) name or position of the second variable (y-axis) to be plotted in environmental space. It must be different from the first one. Default = NULL, required when
selection_type= "random" or "G".- selection_number
(numeric) number of selection to be plotted. Default = 1.
- region_border
(logical) whether to add region border to the plot. Default = TRUE.
- mask_border
(logical) whether to add mask border to the plot. Ignored if mask is not present in
master_selection. Default = FALSE.- col_all
colors for points in all points in the region of interest. The default, NULL, uses a light gray color.
- col_sites
color for selected sites. The default, NULL, uses a blue color to represent selected sites.
- col_pre
color for preselected sites. The default, NULL, uses a red color to represent preselected sites. Ignored if preselected sites are not present in
master_selection.- cex_all
(numeric) value defining magnification of all points relative to the default. Default = 0.7.
- cex_sites
(numeric) value defining magnification of selected sites relative to the default. Default = 1.
- cex_pre
(numeric) value defining magnification of preselected sites relative to the default. Default = 1. Ignored if preselected sites are not present in
master_selection.- pch_all
(numeric) integer specifying a symbol when plotting all points. Default = 16.
- pch_sites
(numeric) integer specifying a symbol when plotting points of selected sites. Default = 16.
- pch_pre
(numeric) integer specifying a symbol when plotting points of preselected sites. Default = 16. Ignored if preselected sites are not present in
master_selection.- add_main
(logical) whether or not to add fixed titles to the plot. Default = TRUE. Titles added are "Environmental space" and "Geographic space".
- mar
(numeric) vector of length 4 to set the margins of the plot in G. The default, NULL, is (3.1, 3.1, 2.1, 2.1) for
plot_sites_Gand (3.5, 0.5, 0.5, 0.5) forplot_sites_EG.- main
(character) the main title for the plot.
- xlab
(character) label for the x axis. The default, NULL, uses variable_1.
- ylab
(character) label for the y axis. The default, NULL, uses variable_2.
Value
plot_sites_EG returns a two-panel plot showing the selected sites.
They are show in both spaces, geographic and environmental.
plot_sites_E returns a plot of selected sites in environmental space.
plot_sites_G returns a plot of selected sites in geographic space.
Examples
# Data
m_selection <- read_master(system.file("extdata/m_selection.rds",
package = "biosurvey"))
# Plotting
plot_sites_EG(m_selection, selection_type = "E")
 plot_sites_E(m_selection, selection_type = "E")
plot_sites_E(m_selection, selection_type = "E")
 plot_sites_G(m_selection, selection_type = "E")
plot_sites_G(m_selection, selection_type = "E")